The name of our first-of-kind (software-based) technology to measure simultaneously the quantitative activity of 20 essential signal transduction pathways in any cell or tissue sample is called: Simultaneous Transcriptome-based Activation Profiling (STAP) Signal Transduction Pathway (STP) technology, in brief: STAP-STP. This technology enables precision therapy development in principle for any disease, based on unique mechanistic insight. Our proprietary approach is expected to increase chances at success in clinical studies, and decrease time to regulatory approval.
In Metamorphosis Tx, the subsidiary of DCDC Tx with BioDetection Systems as co-founder, the STAP-STP technology has been complemented by the CALUX-STP technology of BioDetection Systems.
Therapy development within DCDC Tx aims at complex diseases with a high medical need, such as Post-COVID and sepsis. R&D is dominantly financed by public-private grants with ‘in kind’ contribution of STAP-STP technology.
Metamorphosis Tx, the subsidiary of DCDC Tx, focuses on development of potentially curative cancer differentiation therapy for the most aggressive cancer types. Preliminary results are available for glioblastoma, breast and ovarian cancer (e.g. collaboration with Stanford University). These cancers are the initial focus of Metamorphosis.
STAP-STP Technology
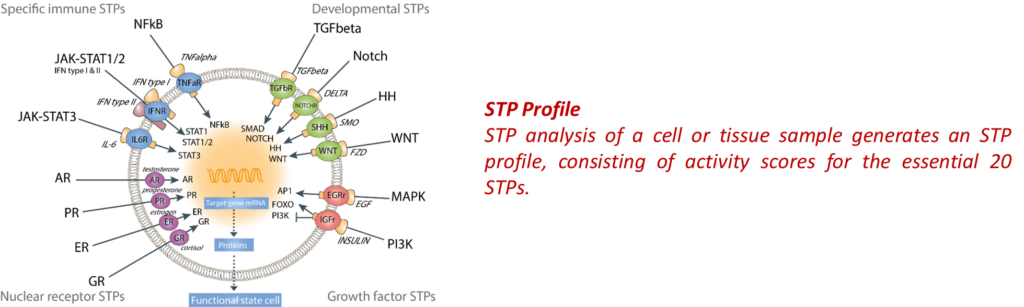
Signal Transduction Pathways (STPs) are the biological processes that regulate functions of all cell types in the body, and a change in STP activity is directly related to a change in cell function.

STAP-STP technology (software) enables quantitative measurement of activity of the 20 relevant STPs simultaneously on any cell or tissue sample (from the laboratory or patient), based on transcriptome (RNA) analysis. This results in a series of STP activity scores, together called the STP profile.
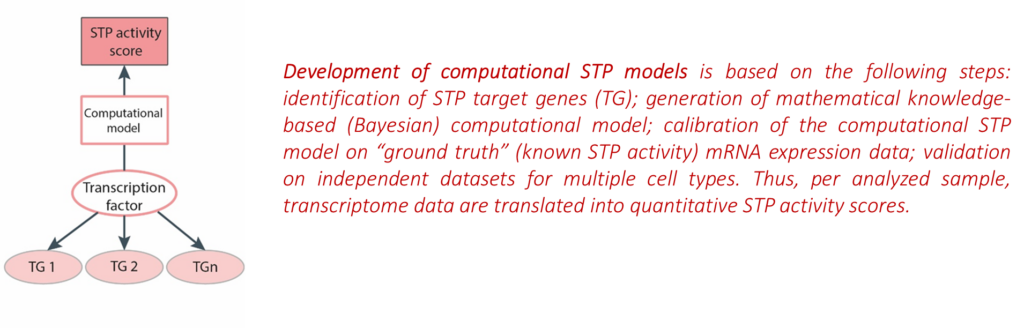
STAP-STP technology is based on artificial intelligence: probabilistic (Bayesian) mathematical modeling is used to quantitatively infer activity of an STP from measured mRNA concentrations of target genes of the transcription factor of an STP.
STAP-STP technology (software) can be used to analyze (transcriptome) data of thousands of preclinical and clinical studies in public databases, such as GEO. This provides completely unique information on the mechanism of disease, mechanism of action of drugs, laboratory disease models etc.. This forms the ‘in silico’ basis for the therapy development program.
Therapy development within DCDC Tx is primarily focused on drug repurposing, however new drug development is in scope.
Scientific approach
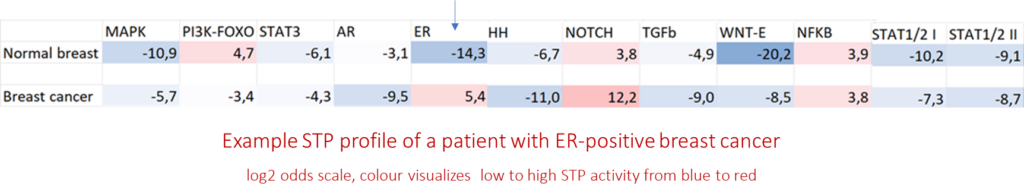
In a cell or tissue sample the clinically relevant STPs are measured, resulting in a series of STP activity scores, together called the STP profile. By comparing STP-profiles between healthy individuals and patients, abnormal STP activity causing abnormal functioning of diseased cells, is uncovered. A disease-associated change in the STP profile is defined as the ‘mechanism of disease’. Currently, making use of transcriptome data in public databases, over 150 diseases have been investigated using STAP-STP technology. In all investigated diseases abnormal activity of specific STPs was discovered as the ‘mechanism of disease’. Except for a number of cancer types, such as breast and prostate cancer, no information had been previously available on STP activity in human disease samples. Thus, by using STP technology to analyze human sample RNA data, new mechanisms of disease were uncovered, or for the first time identified in human patients.
To develop new therapies for diseases, DCDC-Tx investigates in cell-based disease models in the lab which drugs turn the abnormal STP-profile (the ‘mechanism of disease’) into a ‘healthy’ STP-profile. If necessary, a test (companion diagnostic test) is developed to select the subset of patients expected to benefit from the developed therapy. This is important, given that nowadays most patients with the same clinical diagnosis receive a similar therapy, which is often found to be only effective in a subset of patients, due to unrecognized differences in the underlying ‘mechanism of disease’3.
- STAP-STP technology is based on artificial intelligence: probabilistic (Bayesian) mathematical modeling is used to quantitatively infer activity of an STP from measured mRNA concentrations of target genes of the transcription factor of an STP. STAP-STP technology can be distinguished from conventionally used bioinformatics methods for RNA analysis by: (1) excellent biological validation (30 peer-reviewed publications); (2) focus on disease mechanism; (3) suited for analysis of an individual sample, all cell types; (4) suitable for use as companion diagnostic for personalized medicine.
- Conventionally used RNA-analysis methods, such as Gene Set Enrichment Analysis (GSEA), are used for biomarker discovery, including drug targets, based on comparison of multiple groups of sample data to discover statistically relevant correlations (no causal relations) with molecular biomarkers. STAP-STP technology is not used for molecular drug target discovery. More details on the comparison between STP analysis and conventional data analysis tools can be found in the STAP-STP technology publications.
- A treatment is usually effective for a subset of patients with a specific disease. With STAP-STP technology over 150 diseases were characterized, and frequently variations in the mechanism of disease were discovered, fully in line with clinical experience (see STP technology publication list).
